Research
Our laboratory investigates genetics and epigenetics underlying the evolution of genes and genomes in plants, especially origination and functionalization of the genomic novelties. Our approach to these problems will be combining our investigations in the model organism Arabidopsis and the critical crop plant species Oryza. We will focused on the three major directions to understand the exciting process leading to fixation and phenotypic evolution of evolutionary novelties, especially the newly evolving genes in recent evolutionary times.
I. New gene functionality in Arabidopsis
We will use comparative genomic analysis in A. thaliana and its closely related species to identified new gene duplicates which originated recently in these species (<10 million years old), and to characterize the functionality and evolutionary patterns of these new genes.


We will first characterize the gene structures of these new genes using next generation sequencing and computational analysis of public genome sequence data in some of the Arabidopsis species. We will conduct the sequence evolutionary analysis using the current models to test the evolutionary forces of natural selection, genetic drift and mutations.
We will investigate the polymorphic duplicates in Arabidopsis by using the next generation sequencing to investigate the fixation of new gene duplicates in natural populations of Arabidopsis, and to answer a basic problem to the genome evolution: what is the first step of new gene evolution in plants?
We will also further study the functionality and phenotypic changes of young genes in A. thaliana.
II. Origination and evolution of genes and genomes in rice
Cultivated rice (Oryza sativa and O. glaberrima) was evolved and domesticated from a rapid artificial selection and then adapted to highly diverse environments. Such rapid domestication and broad adaptation could enhance gene and genome origination and evolution, which were previously demonstrated. However, an outstanding problem remains unclear that how genes with novel functions originate through rice domestication in a very short evolutionary period. The lack of study in this problem hampered our understanding of how rice-specific biology was evolving from, the plant functional genomics plant in general, and finally the improvement of the cultivated rice.
Two levels of questions about this process can be defined. First, at the level of individual new gene, what are the initial molecular mechanisms and driving forces that generate and maintain new gene structures? Second, at the level of genome, how often do new genes originate and are there any patterns that underlie this process. We will use comparative genomics, population genetics, molecular evolution, and molecular biology approaches to systematically detect and characterize the major features of new genes in cultivated rice, O. sativa and O. glaberrima, the independently Asian and African domesticated rice species, respectively. Specific objectives include: This is the body text of the website
- Comparative genomic analysis of whole genomic sequences from genus Oryza to identify new genes recently originated in O. sativa and O. glaberrima.
- Investigation the pattern and mechanism of new genes recently evolving in O. sativa and O. glaberrima. We will approach the evolution and natural selection of newly evolved genes in cultivated rice and their wild relatives using molecular evolution theory and population genetic analysis.
- Detection the functional and phenotypic significance of new genes in O. sativa. I will conduct two levels of functional analyses. The short-term goal is to compare the expression profiles and/or transcriptom data between novel genes and their parental counterparts. The long-term goal is to use molecular genetics approaches to observe morphological, developmental, and physiological changes.
- Comparison of new genes and domestication genes between O. sativa and O. glaberrima: comparison of new genes independently evolved from each species; the evolution pattern of domestication genes in O. glaberrima.
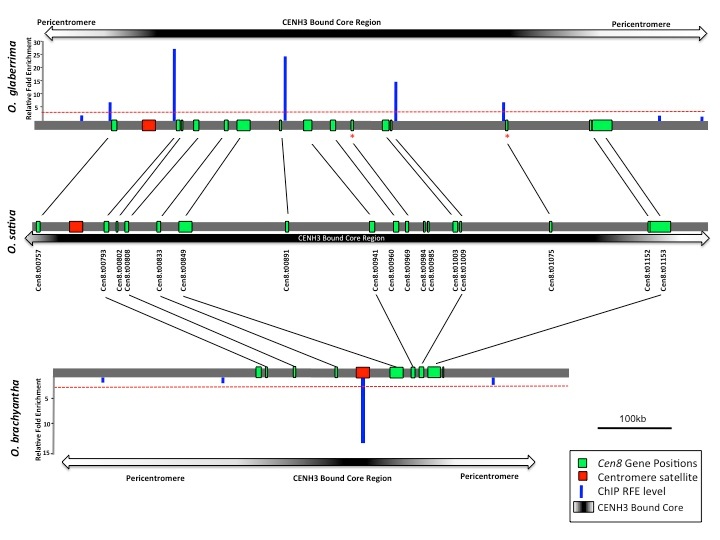
III. Evolution and biological function of centromeric genes
Centromeres, which are defined by the presence of a centromere-specific histone variant (CENH3) and characterized by repetitive DNA, are completely devoid recombination by chromosomal crossing over. Molecular population genetics and molecular evolutionary theories predict that, in chromosomal regions with suppressed or reduced recombination, deleterious mutations can more easily accumulate due to inefficient natural selection, which is thought to be a major factor leading to genetic degeneration of genes and genomes. We will extend the theoretical prediction and the experimental analysis to the more general genomic regions of the suppressed recombination: the centromere of chromosome 8 (Cen8) in the rice genomes, which contains 16 transcribed genes. We develop a set of specific research objectives of centromeric gene evolution. (1) Origination of conserved Cen8 gene. (2) Evolution pattern of Cen8 conserved gene using population genetic analysis. (3) Biological function of Cen8 genes in rice.